[/vc_column_text][/vc_column][/vc_row]
Mechanisms of resistance
Resistance to macrolides occurs by three main mechanisms: target modification, enzymatic drug inactivation, and active efflux. (5,6)
Target modification
Methylation of the binding site in the ribosome is the most prevalent of the resistance mechanisms and confers resistance to both macrolides and lincosamides by preventing their binding to their target sites.(7,8)
Methylation is carried out by enzymes (methylases) encoded by different erm (erythromycin ribosome methylase) genes.
The expression of the methylases is normally inhibited by the ability of the encoding mRNA to adopt a conformation that blocks the initiation of the translation. In the presence of an inducer (macrolide) and its binding to the ribosome, the mRNA re-arranges unmasking the initiation sequences for the translation of the methylase allowing for its expression and conferring resistance to macrolides (Figure 1). (7,9,10)
A
B
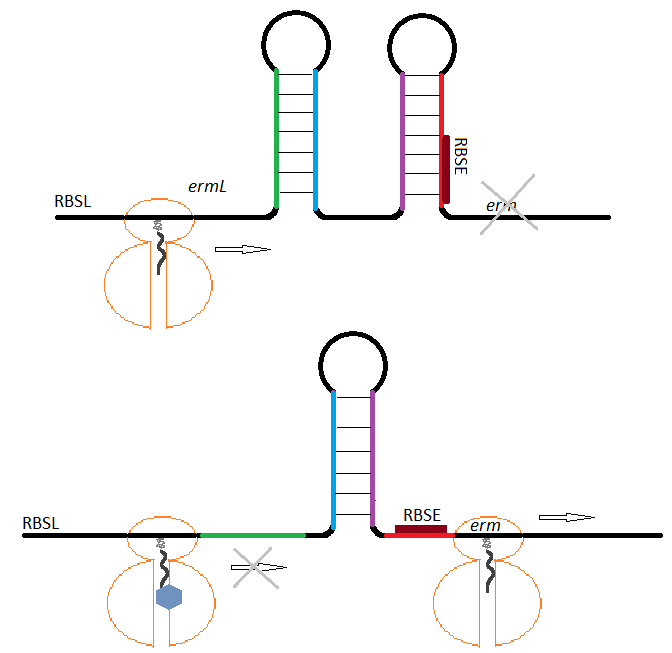
Figure 1. Schematic representation of erm expression regulation. In the absence of an inducer (A), the methylase gene erm is not translated because its ribosome binding site (RBSE) is sequestered in mRNA secondary structure. During induction (B), an erythromycin-bound ribosome stalls at the ermL leading to a change in the mRNA conformation allowing translation of erm.




